|
NOVIDADES
“Soft matter”—those materials that easily deform or structurally change with thermal fluctuations or weak forces—often have the ability to spontaneously form complex structures through the self-organization of molecules. Everyday items such as soaps and detergents and liquid crystal displays in electronic devices are made of these materials. But it is nature that has really perfected the art of molecular self-assembly. Biological molecules are a type of soft matter that are particularly promising for the development of new materials and devices. Dmytro Nykypanchuk—a materials scientist in the Soft and Bio Nanomaterials Group at the Center for Functional Nanomaterials (CFN), a U.S. Department of Energy (DOE) Office of Science User Facility at Brookhaven National Laboratory—has been investigating how the biomolecule deoxyribonucleic acid (DNA) can be used to guide the self-assembly of materials at the nanoscale. In particular, he is studying organic-inorganic hybrid nanomaterials whose structures evolve in response to environmental cues or external stimuli. His work has relevance to drug delivery, sensing, energy, electronics, and other applications.  In an x-ray lab at the Center for Functional Nanomaterials (CFN), materials scientist Dmytro Nykypanchuk sets up a measurement on a small-angle x-ray scattering instrument to determine the nanostructure of a polymer sample.
For example, soap molecules in water form spherical structures called micelles. But this simplistic self-assembly only goes so far in creating specific structures because the molecules can only combine according to their innate intermolecular interactions. In order to expand self-assembly to a much wider range of structures and material types, we need to think first about how to decouple these interactions. In other words, the material you are assembling should not define the structure itself. Or, the brick should not define the building; you should be able to build whatever building you want from that brick. 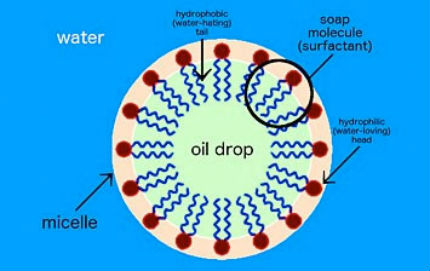 Soap is a surfactant, or surface-active agent. One end of the soap molecule is hydrophilic (water-loving) and thus binds to water; the other end is hydrophobic (water-hating) and binds to oil molecules. In water, soap molecules form a ring around the drop of oil. This structure is called a micelle.
Watson and Crick discovered the double-helix structure of DNA in 1953, and that discovery has led to much research into how genes control the chemical processes within cells. This different application of DNA arose from scientists trying to get inspiration from nature. Nature can create very complex designs that are autonomous and functional—like the human body, for example. However, DNA is not the only information carrier in nature. Other scientists are studying how biomolecules like peptides, proteins, and RNA could be used as programmable “smart” building blocks for self-assembly. Each of these approaches has advantages or disadvantages and, depending on the application, the use of peptides versus DNA could be more advantageous, or vice versa. Biologically inspired self-assembly holds the most promise compared to other self-assembly methods, but at the same time, there are numerous technical difficulties and limitations. One of the most obvious limitations is that biologically inspired self-assembly is typically limited to aqueous solutions and to near-room temperatures. A simpler approach relies completely on nonspecific interactions between molecules. Block copolymers are an example of this kind of self-assembly, where one molecule has two chemically distinct components. The interaction of these components leads to microphase separation at the nanoscale and ultimately to the creation of a variety of ordered nanostructures. In attempt to widen the use of nonspecific self-assembly, scientists also explore directed self-assembly approaches, in which the materials assemble according to their combined innate interactions, but the assembly is guided by externally applied thermal, optical, electrical, or chemical fields or gradients. For example, one of the groups at CFN uses laser light to drive self-assembly. 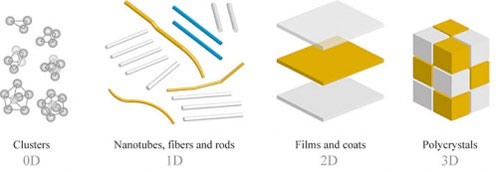 Nanostructures can exist in zero, one, two, or three dimensions.
With 2D assembly, we have more methods available to visualize the complete structures and all the internal components, such as atomic force microscopy and scanning and transmission electron microscopy. 2D assembly also makes it easier to explain or describe the formation mechanisms or structural changes that occur with changing environments. The advantages of material dimensionality depend on the application. For example, today’s electronic technology is mostly 2D. Computer chips are based on 2D thin films. Fabrication of 1D systems like nanowires can be used to build out nanocomponents, which are then assembled into circuits for electrical, optical, or chemical signals. Cryo-electron microscopy (cryo-EM) is another imaging technique that our group has started to use more often. In this technique, electrons are directed toward samples that have been frozen in solution and preserved in their native configuration. 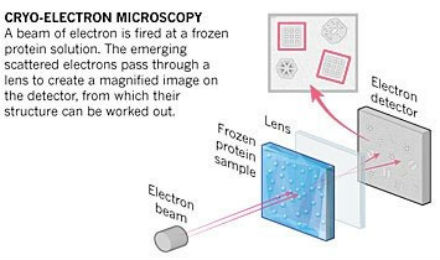 A schematic of cryo-electron microscopy.
This structural flexibility is attractive for applications including drug delivery and sensing. It also is relevant to creating nanoscale “machines” that respond to some change in the environment and do work or convert energy to different forms, such as converting light energy into chemical energy. Ultimately, we want to be able to have absolute control over the dimensions, configurations, and properties of materials—going from a manufacturing process to creating things on their own by mixing the components necessary to produce the desired structures and functions. The projects are not always directly related to my background and expertise, so I am always learning something new. Interacting with users gives me ideas for my own work and makes me see new applications for how I can apply my expertise in a different direction. For example, I was discussing a project with a user who was synthesizing and studying upconverting nanoparticles, which are nanoparticles that absorb two photons (light particles) and emit a single photon of higher energy—a useful property for many applications, including bio-imaging, bio-sensing, and photocatalysis. We realized that DNA directed self-assembly can be used to address the problem of low upconverting efficiency of the nanoparticles by coupling them with plasmonic particles, typically noble metal particles, via DNA linkage. Using DNA provides a means of precisely controlling the distance between the particles, which is essential for plasmon-enhanced upconversion. 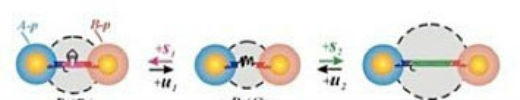 DNA that is linking nanoparticles responds to the addition of molecular stimuli and changes the distance between nanoparticles with nanometer precision.
The CFN also has a strong sense of community. For the past three years during Christmastime at the CFN, we have had a potluck lunch at which everyone brings a dish to be shared. Usually people bring culturally diverse foods, and the lunch provides a really nice time to experience different countries. I am hoping this event will become a CFN tradition! We also have weekly informal gatherings in the lobby to catch up with colleagues and hear about their latest projects. One of my first projects as an undergrad was calculating interactions between particles—in particular, the effect of van der Waals forces on the aggregation of particles, depending on particle size and aggregate configuration. The van der Waals interactions alone are not that exciting for relatively simple cases of one-component systems. But once you start increasing the system complexity, going from single to multicomponent systems, more interesting interactions emerge, and you realize that many processes and technologies in the food industry, environmental science and remediation, and medicine are defined by these interactions. Here is where my interest in DNA fits in. As an information carrier, DNA can be used to program our own “designer” interactions to guide the assembly of particles into “smart” materials—those with properties that can be controllably altered in response to external stimuli. 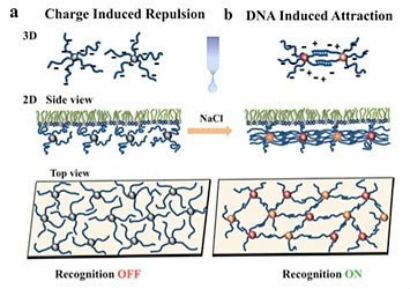 Nykypanchuk and colleagues from the CFN and National Synchrotron Light Source demonstrated a framework for the assembly and tuning of 2D DNA-based nanoparticle systems at liquid interfaces. They tuned the interactions by changing the concentration of salt (NaCl). Without salt, the interactions are dominated by electrostatic repulsion between DNA chains (a). Introducing salt causes an attractive interaction between the nanoparticles because complementary strands of DNA join together.
Because of safety and training regulations, elementary, middle, and high-school students cannot enter the facilities at the CFN. We came up with the idea for a portable videoconferencing system that would allow the students to remotely “enter” the labs and watch as one of their teachers works alongside a Brookhaven scientist to look at nanoparticles. I knew this project was complementary to my expertise, so I wanted to help out. I continue to work with Freeport teachers today. I also participate in a couple of workshops organized by Brookhaven’s Office of Educational Programs (OEP) and NSLS-II. At these workshops, I introduce the CFN facilities and explain to teachers and students how the schools can use them for student projects, with the setup from the Freeport collaboration as a template. To see the enthusiasm of both students and teachers is a very rewarding experience, which further motivates me in my work. I hope that the experiments that students participate in motivate them to learn more about the world around them. Brookhaven National Laboratory. Posted: Sep 17, 2018. |
|||||||||||||||||||||||||